A quick search of blueberry with the terms “anthocyanin and chlorogenic acid” bring up numerous articles extolling the virtues of blueberries as a healthy superfruit. Answering questions about what genetic mechanisms are controlling the accumulation of these bioactive compounds—and their relationship with fruit quality traits—is vital for the improvement of blueberry cultivars.
For instance, a recent study in blueberry indicated that through digestion, acylated anthocyanin (a form of anthocyanin that accumulates in blueberry fruits), are recovered at a higher rate. As a result, the adsorption of these health-related bioactive compounds in the human body could increase.
Recent developments and adoption of advanced genotyping platforms and genomic recourses have advanced our understanding of what controls accumulation of these bioactive compounds in Vaccinium crops. Two recent studies focused on dissecting the genetic mechanisms controlling chlorogenic acid (CHA) and anthocyanin content and their composition in blueberries (Mengist et al., 2022; Montanari et al., 2022).
Study 1. Dissecting the genetic basis of bioactive metabolites and fruit quality traits in blueberries (Vaccinium corymbosum L.)
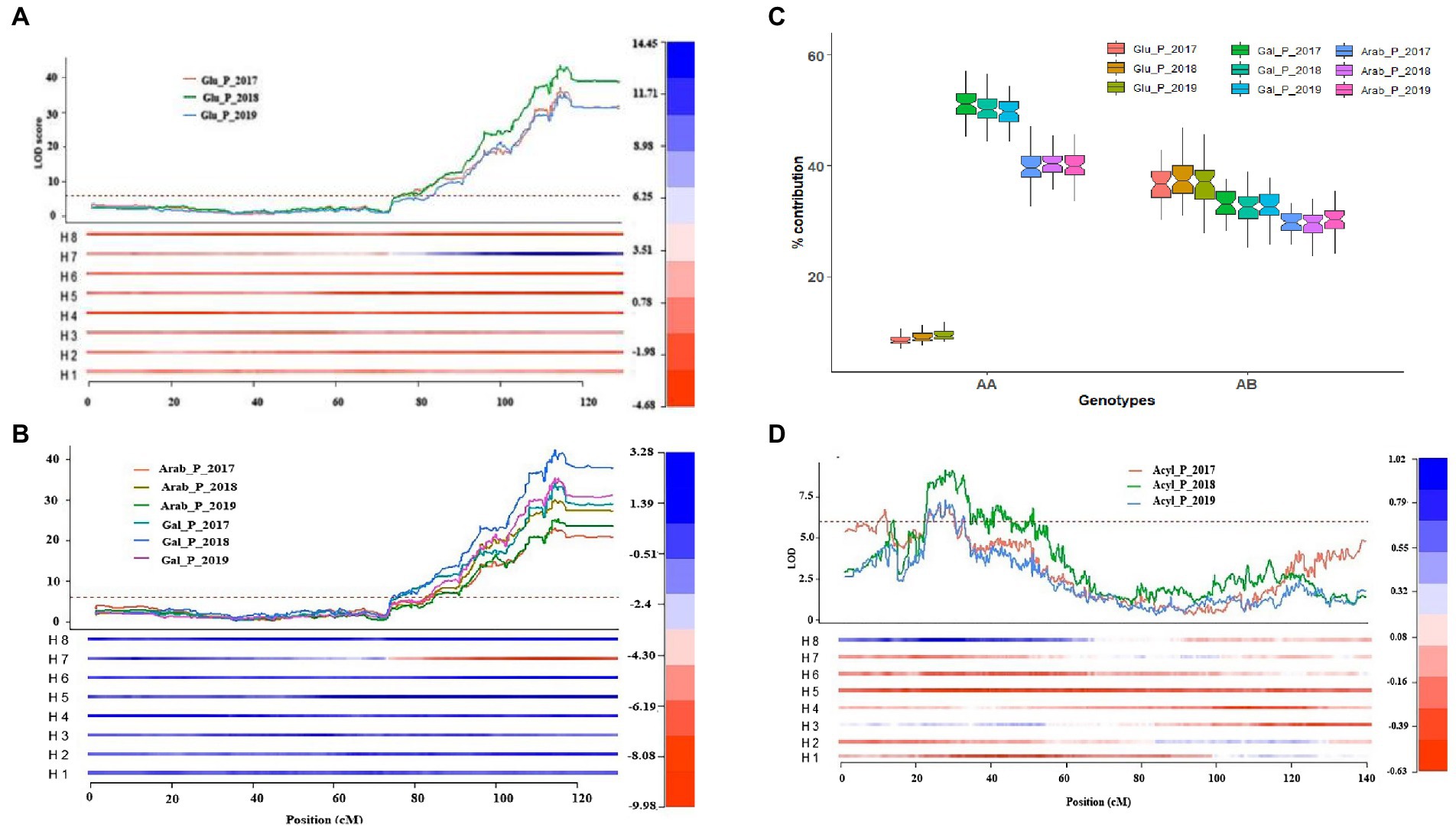
Mengist et al. (2022) used a mapping population (DSxJ) representing highbush blueberry cultivars Draper and Jewel and identified 180 quantitative trait loci (QTLs) for total and individual anthocyanin content, relative anthocyanin composition, and CHA.
A QTL influencing CHA concentration was identified on chromosome 2 that explained up to 21% of the phenotypic variation and was consistent over three consecutive years of the study. This is the only known QTL associated with CHS.
One cluster of stable QTLs influencing total anthocyanin content was mapped on chromosome 1 and explained up to 24% of the phenotypic variation. Four clusters of QTLs controlling the conjugations of anthocyanin with different sugar moieties and acylation were mapped on chromosomes 1, 2, 4 and 8 and were stable across three years.
- The cluster of QTLs that mapped on chromosome 4 explained up to 80% of the phenotypic variance, and was associated with an increase of glucoside-based anthocyanins relative to the arabinoside/galactoside-based anthocyanins.
- A second cluster of QTLs mapped on chromosome 1, explained up to 35% of the phenotypic variance and was associated with reducing the concentration of arabinoside- and galactoside-containing anthocyanins.
- A third cluster of overlapping QTLs mapped on chromosome 8 associated with reducing cyanidin-galactoside and peonidin-galactoside and increasing cyanidin-arabinoside concentration, and explained up to 44% of phenotypic variance.
- A cluster of stable QTLs mapped on chromosome 2 controlled content or proportion of acylated anthocyanin, explained up to 27% of the phenotypic variance and overlapped with the CHA QTL.
Haplotype analysis indicated that the QTLs for CHA and acylated anthocyanin were in different haplotypes, which implies they could be selected independently.
Study 2. High-density linkage map construction in an autotetraploid blueberry population and detection of quantitative trait loci for anthocyanin content

Montanari et al. (2022) used recently released tools for genetic mapping to build a high-density linkage map and to detect QTLs for fruit anthocyanin content. The team used a different mapping population (NxHB) representing two different highbush cultivars, Nui and Hortblue Petite.
- They identified two clusters of stable QTLs on chromosomes 2 and 4, which explained up to 77.5% and 39.0% of the phenotypic variation for anthocyanin composition, respectively.
- QTLs mapped in chromosome 4 had a positive effect on the concentrations of most of the glucoside based-anthocyanins, and a negative effect on most of the arabinoside/galactoside-based anthocyanins.
Integration of results from the two studies highlighted that:
- The two clusters of QTLs controlling acylation and glycosylation mapped on chromosomes 2 and 4 across the two studies were anchored to the W85 v2 genome (Mengist et al., 2022); this revealed that they overlap.
- The QTLs on chromosome 2 overlap in a 3.2 Mb region spanning position 8 to 11.2 Mb, and the QTLs in chromosome 4 overlap in a 4.8 Mb region spanning position 56.2 to 61Mb.
- Overlapping of these QTLs across different genetic backgrounds represents the first level of QTL validation and allowed for identification of possible genes controlling anthocyanin acylation and glycosylation.
- Across the two studies several minor-effect and non-stable QTLs were also identified, indicating that environmental factors affect anthocyanin accumulation.
Overall, these studies provide important insight and pave the way to understanding the genetic mechanisms that control CHA and anthocyanin content and composition in blueberries. Future studies can use these findings to further develop DNA markers to select blueberry cultivars with higher content of these bioactive compounds or for specific anthocyanin composition.
Citations
- Mengist MF, Grace MH, Mackey T, Munoz B, Pucker B, Bassil N, Luby C, Ferruzzi M, Lila MA and Iorizzo M. 2022. Dissecting the genetic basis of bioactive metabolites and fruit quality traits in blueberries (Vaccinium corymbosum L.). Front. Plant Sci. 13:964656. doi: 10.3389/fpls.2022.964656
- Montanari S, Thomson S, Cordiner S, Günther CS, Miller P, Deng CH, McGhie T, Knäbel M, Foster T, Turner J, Chagné D and Espley R. 2022. High-density linkage map construction in an autotetraploid blueberry population and detection of quantitative trait loci for anthocyanin content. Front. Plant Sci. 13:965397. doi: 10.3389/fpls.2022.965397













