Investigating chromosomal recombination—the exchanging of genetic material between chromosomes—behavior is crucial to transfer traits under genetic control from one plant to another one and for advancing genetic discoveries. Factors that can limit chromosome recombination include preferential pairing in the case of polyploid species and chromosome rearrangements (e.g., chromosome inversions or translocations). Understanding chromosome genetic behavior and structure of blueberry chromosomes is relevant for two reasons:
- Highbush blueberry is a polyploid species carrying four copies of each homologous chromosomes.
- The genome of blueberry cultivars is heterogeneous, harboring pieces of DNA from wild diploid species that are used in breeding programs to introgress favorable traits (e.g., low chilling requirement).
Due to these characteristics, the blueberry genome is complex and it remains unclear if it behaves as an autopolyploid where each of the four homologous chromosomes recombine randomly or if they recombine preferentially in pairs of two like in allopolyploid plants. It is also still unknown if its genome harbors any chromosome rearrangements that can affect recombination.
In this study, a high-quality reference genome and three linkage maps were developed along with three available genomes representing the cultivar Draper, and two wild species (V. darrowii and V. mirtillus). These resources were used to compare the structure of the genomes, and to assess chromosome recombination behavior. Several methods were integrated into this study.
Plant material, sequencing, and genome assembly
The genome assembly was developed using a wild diploid species V. caesariense clone W85, also known as diploid blueberry (V. corymbosum) and a potential progenitor of tetraploid cultivated blueberry. Assembly of the W85 genome was performed using long reads sequencing technology (PacBio) along with integration of a linkage map, including 17 ,486 single-nucleotide polymorphism (SNP) markers (Qi et al., 2021), that was used to anchor the genome.
Gene prediction and annotation
Gene prediction was performed using Maker v.3.01.03 (Cantarel et al., 2008) by integrating ab initio gene prediction and evidence-based prediction and was performed independently on the two haplotypes.
Repetitive sequence annotation and analysis
One million random read pairs of W85 (NCBI accession no. SRR837868) were analyzed using Tarean (Novák et al., 2017) to identify potential satellite DNA sequences—highly repetitive DNA consisting of short sequences repeated many times.
Selected satellites of W85 were analyzed by fluorescence in situ hybridization (FISH) using young flower buds of the W85, V. darrowii and of the tetraploids ‘Draper’ Selection-44392 , ‘Draper’, and ‘Jewel’ , according to published procedures (Iovene et al., 2008). Oligonucleotide probes and PCR primers were designed using the consensus sequences of the repeats.
Linkage map construction
Three F1 mapping populations named DS × J (n = 196), R × A (n = 346) and D × B (n = 168), were used for linkage map construction. Abbreviations stand for ‘Draper’ (D) and Draper Selection-44392 (DS are northern highbush blueberry (V. corymbosum; NHB) selections/cultivars, whereas Arlen (A), Jewel (J), Biloxi (B) and Reveille (R) are southern highbush cultivars (V. corymbosum interspecific hybrids; SHB) cultivars with 10–60% of their genome represented by wild species.
Estimation of double reduction, quadrivalent, and preferential chromosome pairing
The rate of double reduction (DR) and quadrivalent chromosome pairing were estimated using TetraOrigin software (Zheng et al., 2016). For a given marker, the probability of DR rate was averaged over the number of offspring on parental meiosis. The quadrivalent chromosome pairing was calculated by dividing the number of offspring with the quadrivalent pairing by the total number of offspring in the mapping population.
The meiotic pairing behavior of the 4× genotypes ‘Arlen’, DS, ‘Draper’, and ‘Jewel’ was also evaluated cytologically, by examining at least 40 pollen mother cells at diakinesis-metaphase I in each variety.
Key Results
W85 high-quality genome assembly
The contiguity of the W85 genome assembly was relatively higher than currently available blueberry genome assemblies. In total, 34,895 genes were predicted and about 45% of the genome was annotated as repetitive sequence, which is comparable to the tetraploid ‘Draper’ genome representing similar gene and repeat fractions (32, 139 genes/haplotype, 44.3% repeats) (Colle et al., 2019).
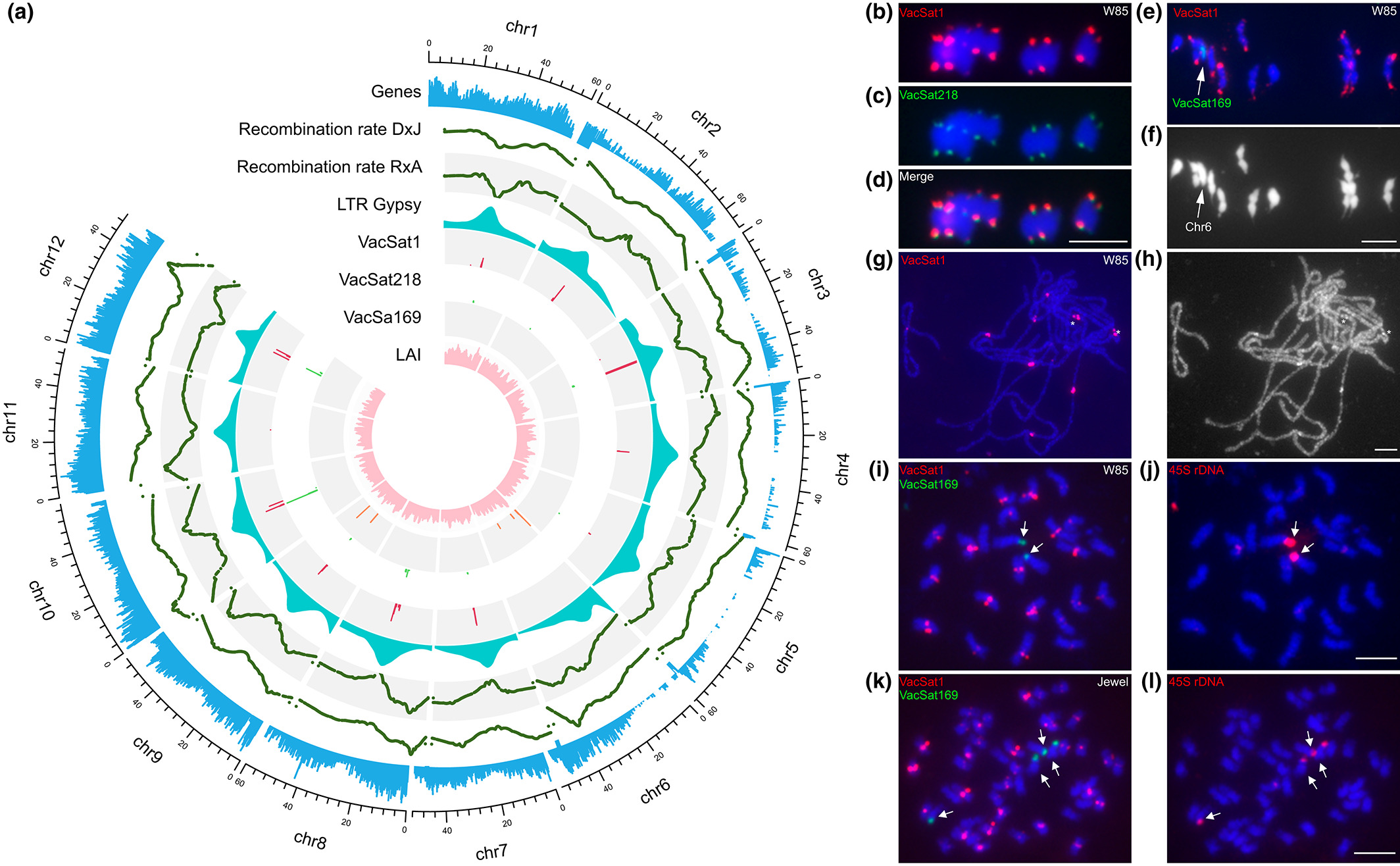
Centromeric repeat VacSat1 is conserved across diploid and tetraploid blueberry species
A putative centromeric repeat (named VacSat1) present in 10-11 chromosomes was identified. VacSat1 sequence and distribution was conserved in tetraploid and diploid species, V. darrowii, and V. mirtillus.
A satellite repeat named VacSat169 was identified on chromosome 6, and was associated with ribosomal DNA, a region known to have low recombination frequency.
Comparative analysis between W85 and tetraploid genomes highlights a reciprocal translocation in the ‘Draper’ genome
A major reciprocal inter-chromosomal translocation was identified between ‘Draper’ chromosome 6 and chromosome 10. The translocation spans the location of VacSat169 in chromosome 6. Reciprocal translocation is a type of chromosomal abnormality that occurs when two non-homologous chromosomes break at the same time, and the broken ends reattach to the opposite chromosome, resulting in a reciprocal exchange of genetic material between the two chromosomes.
The reciprocal translocation was not identified in the other five highbush cultivars and in the wild species.
Besides the chromosome 6/ chromosome 10 translocation the W85 genome was highly collinear with linkage maps, the ‘Draper’, and wild species genomes.
The inter-chromosomal translocation alters chromosome 6 and chromosome 10 meiotic behavior in the ‘Draper’ genome
Analysis of chromosomal recombination in F1 progenies derived from crosses between ‘Draper’; harboring the translocation, and ‘Biloxi’, not harboring the translocation, indicated that the translocation altered chromosome 6 and 10 pairing, recombination, and segregation behavior, and in turn, affected the linkage map construction.
Blueberries behave as an autotetraploid with non-preferential chromosome pairing also called tetrasomic inheritance
DNA Maker analysis indicated that there was no preferential pairing among each of the four homologous chromosomes. Also, there was a substantial quadrivalent chromosome pairing in DS × J and R × A mapping populations, as also indicated by the cytological analysis of three parental varieties at diakinesis-metaphase I (‘Arlen’, DS, and ‘Jewel’).
Overall, our results did not reveal strong enough evidence to demonstrate any preferential pairing in blueberry.
Discussion
High quality genomic resources provide novel insights into the structure of the blueberry genomes
In this study, a high-quality phased assembly of the blueberry genome was released, improving upon previous versions. The genome enables the localization of regions enriched by low complexity sequences (repeats) and highlighted that the structure of the genomes among Vaccinium species (section Cyanococcus) is highly conserved.
Evidence and impact of a reciprocal translocation for blueberry genetic analysis and breeding
This study demonstrated the presence of reciprocal translocation, which formed two fused chromosomes, chromosome 610 and chromosome 106. The co-localization between VacSat1 and VacSat169 that repeats on the same chromosome is specific to the translocation chromosome 106, making these ideal cytogenetic markers for future studies.
The translocation was found to cause multiple abnormal chromosome pairing configurations, which affected recombination between chromosome 6 and chromosome 10. The reciprocal translocation can directly affect phenotypes and cell functionality, such as pollen viability. Also, it can directly affect inheritance of important traits, like cold hardiness, a phenotype that has been associated with a region of chromosome 10. Future work will need to assess the impact of the translocation and evaluate the frequency of the translocation in the blueberry germplasm.
Evidence for autopolyploid genetic behaviors of blueberry
The overall results demonstrated that blueberry behaves as an autopolyploid during meiosis. This implies that the four homologous chromosomes can pair randomly, which increase recombination frequency. This result guides the selection of statistical models to use in genetic studies for blueberry. Models that account for random tetraploid recombination should be used and not those that account for preferential pairing.
Overall, the results of these study provide a framework for comparative genome analysis within Vaccinium spp. and can advance genomic-assisted breeding in blueberry.
Citations
- Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, Holt C, Alvarado AS, Yandell M. 2008. Maker: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Research 18: 188– 196.
- Colle M, Leisner CP, Wai CM, Ou S, Bird KA, Wang J, Wisecaver JH, Yocca AE, Alger EI, Tang H et al. 2019. Haplotype-phased genome and evolution of phytonutrient pathways of tetraploid blueberry. GigaScience 8: giz012.
- Iovene M, Wielgus SM, Simon PW, Buell CR, Jiang J. 2008. Chromatin structure and physical mapping of chromosome 6 of potato and comparative analyses with tomato. Genetics 180: 1307– 1317.
- Novák P, Ávila Robledillo L, Kobližková A, Vrbová I, Neumann P, Macas J. 2017. Tarean: a computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Research 45: e111.
- Qi X, Ogden EL, Bostan H, Sargent DJ, Ward J, Gilbert J, Iorizzo M, Rowland LJ. 2021. High-density linkage map construction and QTL identification in a diploid blueberry mapping population. Frontiers in Plant Science 12: 1– 13.
- Zheng C, Voorrips RE, Jansen J, Hackett CA, Ho J, Bink MCAM. 2016. Probabilistic multilocus haplotype reconstruction in outcrossing tetraploids. Genetics 203: 119– 131.













